Model Summary for yheO
?
Open a dynamic genome browser to visualize ChIP-Seq data and BoltzNet coverage predictions over the full genome
Browser
?
The weight matrix is a json file that include the weight matrix and bias term. The model is a zip file with the tensorflow model directory. Data points to RegulonDB for all ChIP-Seq data
Downloads
?
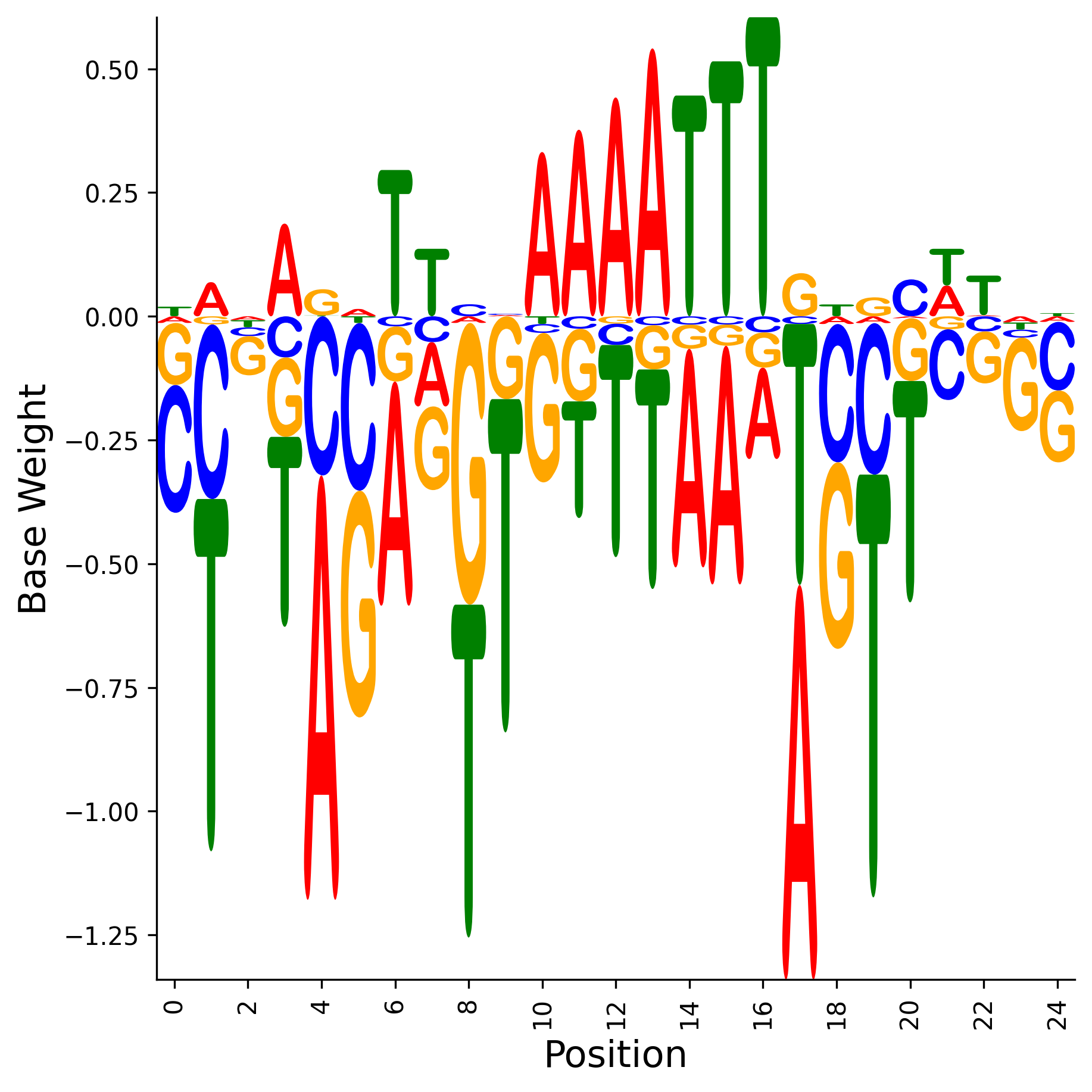
The binding weight matrix (WM)
Weight Matrix
?
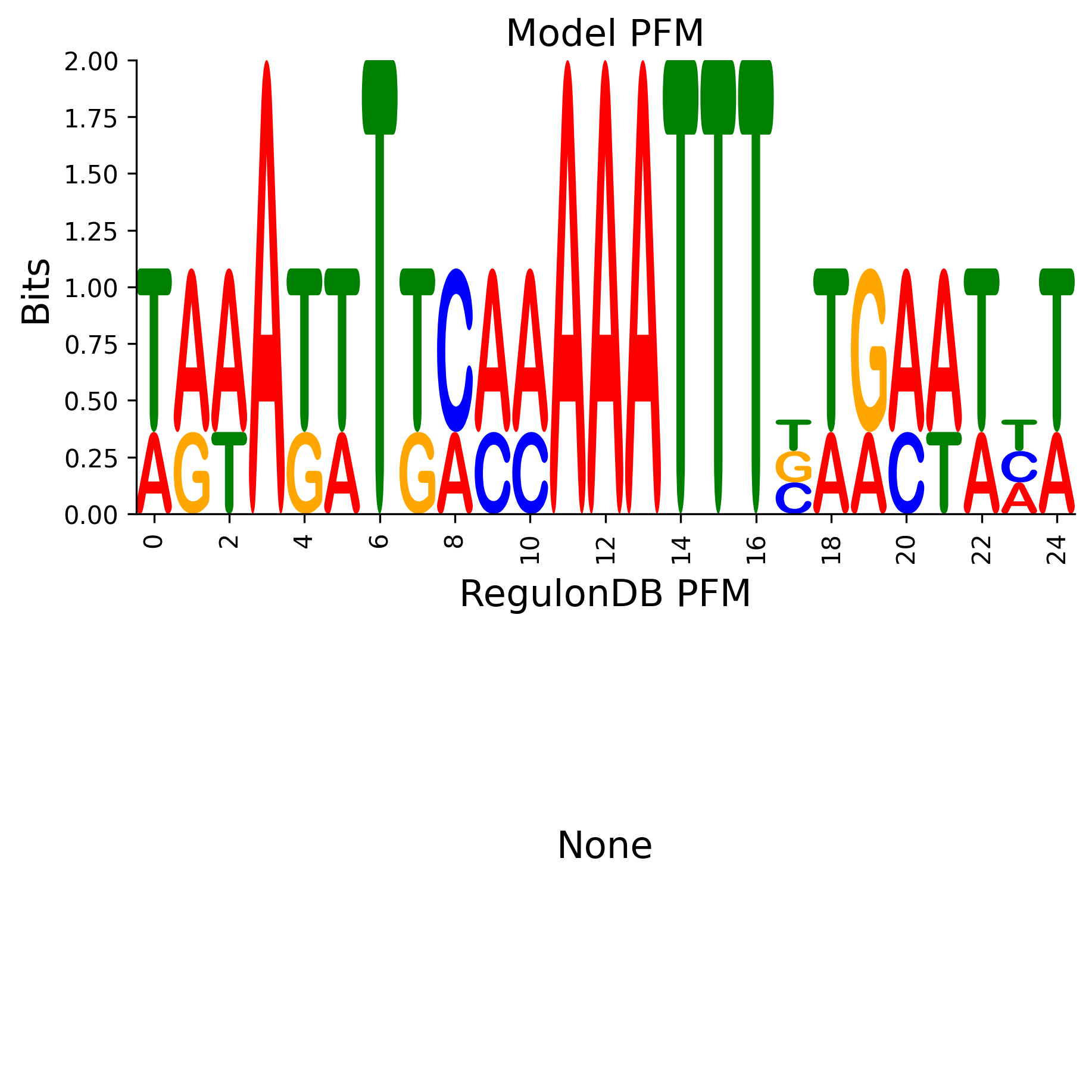
The position frequency matrix (PFM) derived from the weight matrix (WM) and the corresponding RegulonDB PFM if available
Position Frequency Matrix
?
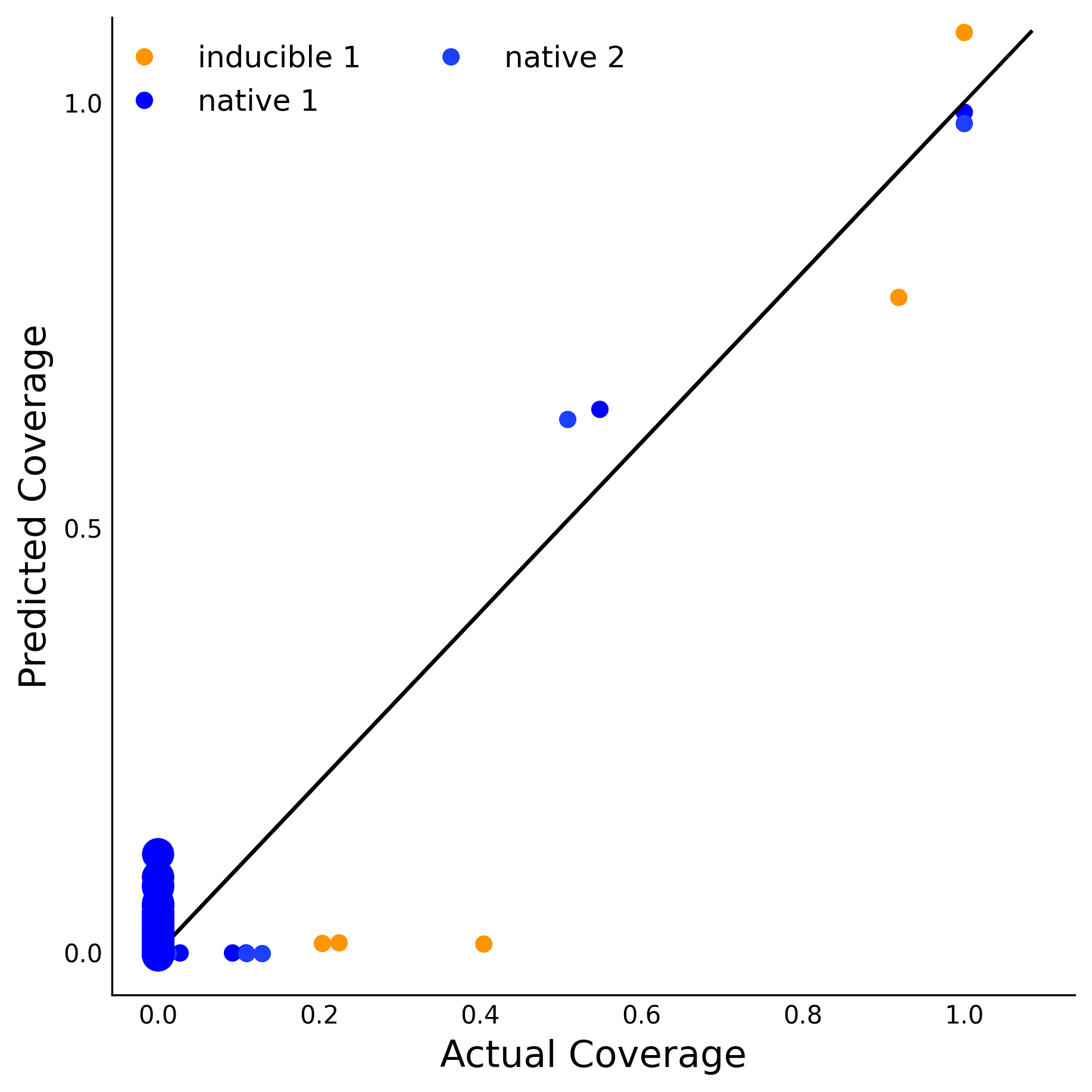
Scatter plot of predicted vs actual ChIP-Seq coverage on training data set in all experiments
Training Data Accuracy
?
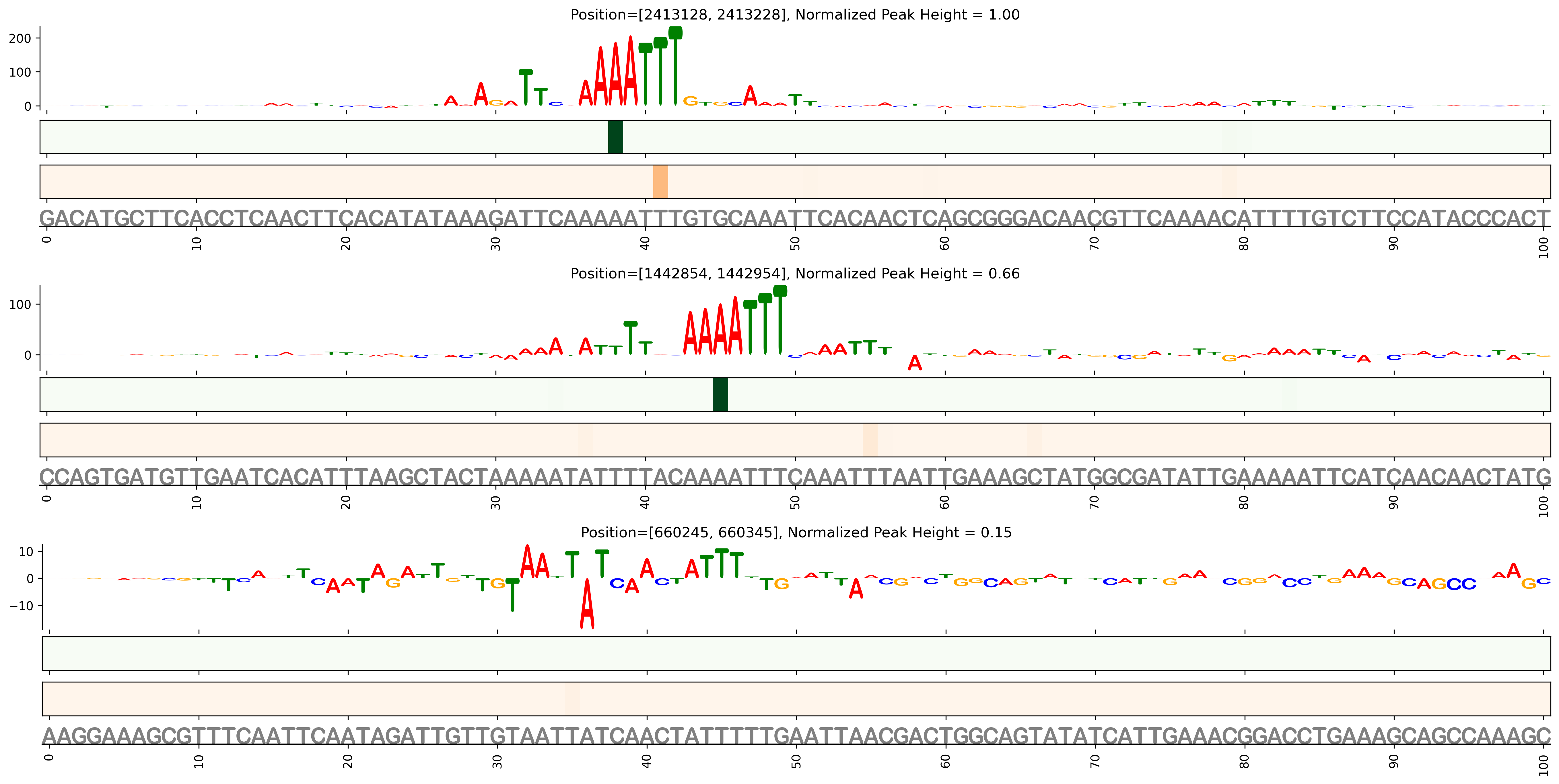
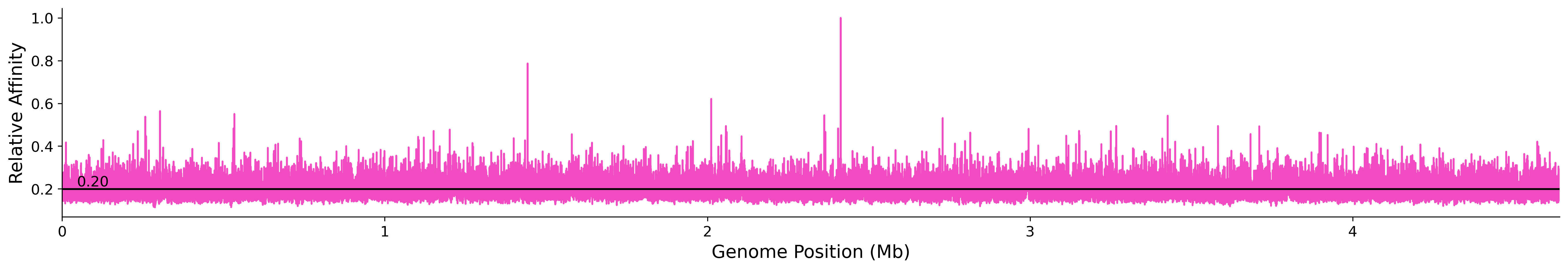
Prediction of binding affinity in 101bp windows over the genome. Affinity scaled to maximum affinity over the genome.
Genome Wide Predicted Affinity
?
Prediction at nucleotide resolution in 101bp windows around top predicted ChIP-Seq binding sites. The logo indicates the contribution of each base to the overall 101 bp sequence affinity (either postive or negative). The heat maps indicate the exponentiated weight matrix score at each postion on the postive (green) or negative (red) strand. The sequence and relative position within the sequence shown at the bottom. Locations of previously reported binding sites shown in cyan box
Predicted Binding in Top ChIP-Seq Binding Sites